SKLB CoralBioinfo Database
Although reef-building corals play an important role in the global marine ecosystem, there emerges a huge lack of bioinformatics data as well as databases from these species, exerting negative effects on exploring their biology. Here we introduce the construction of SKLB CoralBioinfo database series. Developed under the Docker-based LEMP tech-stack, these databases integrate our own data from full-length transcriptome sequencing and small RNA sequencing of several main reef-building coral species with other existing coral bio-data. It is expected for this work to fulfill the “data gap” and boost the virtuous 3A (Acquisition-Analysis-Application) cycle in coral multi-omics research.
SKLB CoralBioinfo Database1. DATABASE CONTENT1.1. Sequence1.2. Gene1.2.1. StructureCDSSSR1.2.2. AnnotationGOKEGGKOGNrNtPfamSwissProt1.3. Other data2. WEB SERVICE3. SUB-DATABASE4. INCLUSION CRITERIA4.1. Genome4.2. Transcript4.3. Small RNA
1. DATABASE CONTENT
CoralBioinfo, which stores basic bioinformatics data such as ▼sequence and ▼gene information, is first developed to lay the foundation for down-stream database construction.
1.1. Sequence
All raw sequences are available on NCBI:
| Species | Full-length transcript | Small RNA | Illumina RNA-seq* |
|---|---|---|---|
| Acropora muricata | SRR9613488 |
SRR13442147 SRR13442146 SRR13442145 |
SRR12904786 SRR12904785 SRR12904784 |
| Montipora foliosa | SRR9129316 |
SRR13442144 SRR13442143 SRR13442142 |
SRR12904783 SRR12904782 SRR12904781 |
| Montipora capricornis | SRR9129315 |
SRR13442141 SRR13442150 SRR13442149 |
SRR12904780 SRR12904792 SRR12904791 |
| Pocillopora verrucosa | SRR9129314 |
SRR13442152 SRR13442151 SRR13442148 |
SRR12904794 SRR12904793 SRR12904787 |
| Pocillopora damicornis | SRR9613489 | NULL |
SRR12904790 SRR12904789 SRR12904788 |
*Illumina RNA-seqs are not stored in CoralBioinfo. However, URLs of their mirrors on public database must be listed according to 4.2.
These raw data are processed through mature bioinformatics pipelines to get Unigene and miRNA sequences, which have been transformed into ncbi-blast databases for alignment tasks.
1.2. Gene
Gene information mined from Unigene is consisted of transcript structure and annotations from seven databases: GO, KEGG, KOG, Nr, Nt, Pfam and Swiss-Prot.
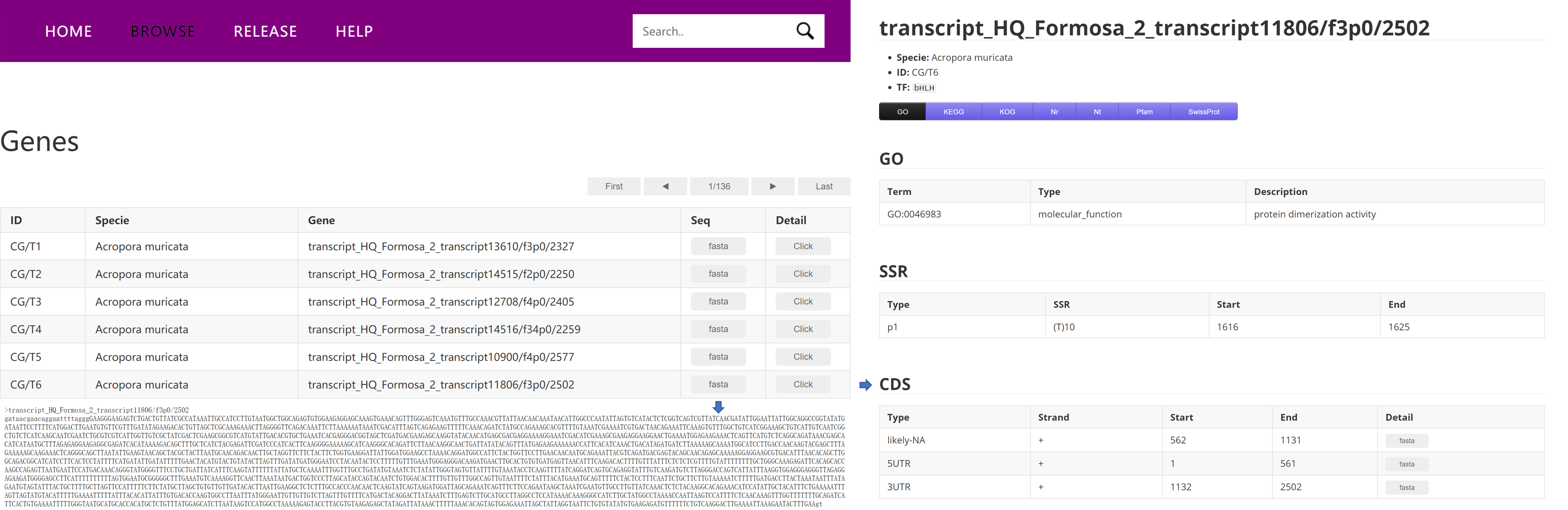
Every gene page demonstrates sequence, annotation and structure information of the transcript. There will be 7 buttons labelled "GO", "KEGG", "KOG", "Nr", "Nt", "Pfam" and "SwissProt" for users to get annotation details. If there are no annotation results for the transcript in one database, corresponding button will be unclickable.
1.2.1. Structure
CDS
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...SH_FW_transcript13494/f2p0/1891 |
| type | <tag>-<completeness> | 3UTR | 5UTR <tag> = confident | likely | suspicious | dumb <completeness> = compl | 5/3partial | internal | NA | confident-compl |
| seq | CDS/UTR sequence | ATGCCGTCGAT... |
| start | start site | 255 |
| end | end site | 782 |
| species | - | Montipora capricornis |
SSR
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...SH_WP_transcript13616/f12p0/2318 |
| type | c (complex repeat) pn (n nt repeat) | p3 |
| SSR | SSR sequence | (ATT)14 |
| start | start site | 1808 |
| end | end site | 1849 |
| species | - | Montipora foliosa |
1.2.2. Annotation
GO
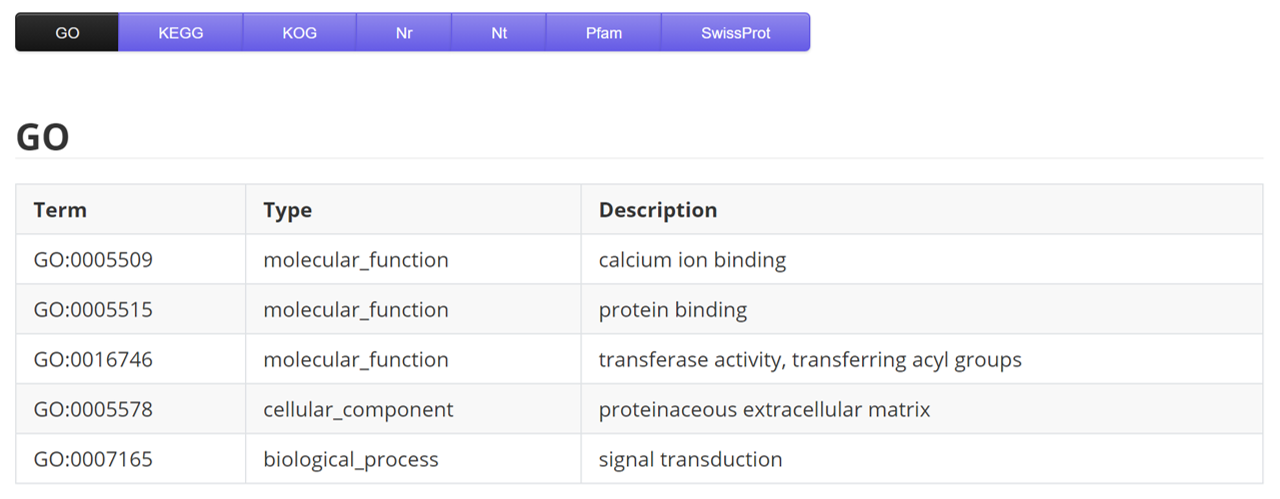
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...HQ_SH_981_transcript2380/f3p0/3329 |
| term | GO term | GO:0007049 |
| type | ontology type | biological_process |
| description | - | cell cycle |
| species | coral species | Pocillopora verrucosa |
KEGG
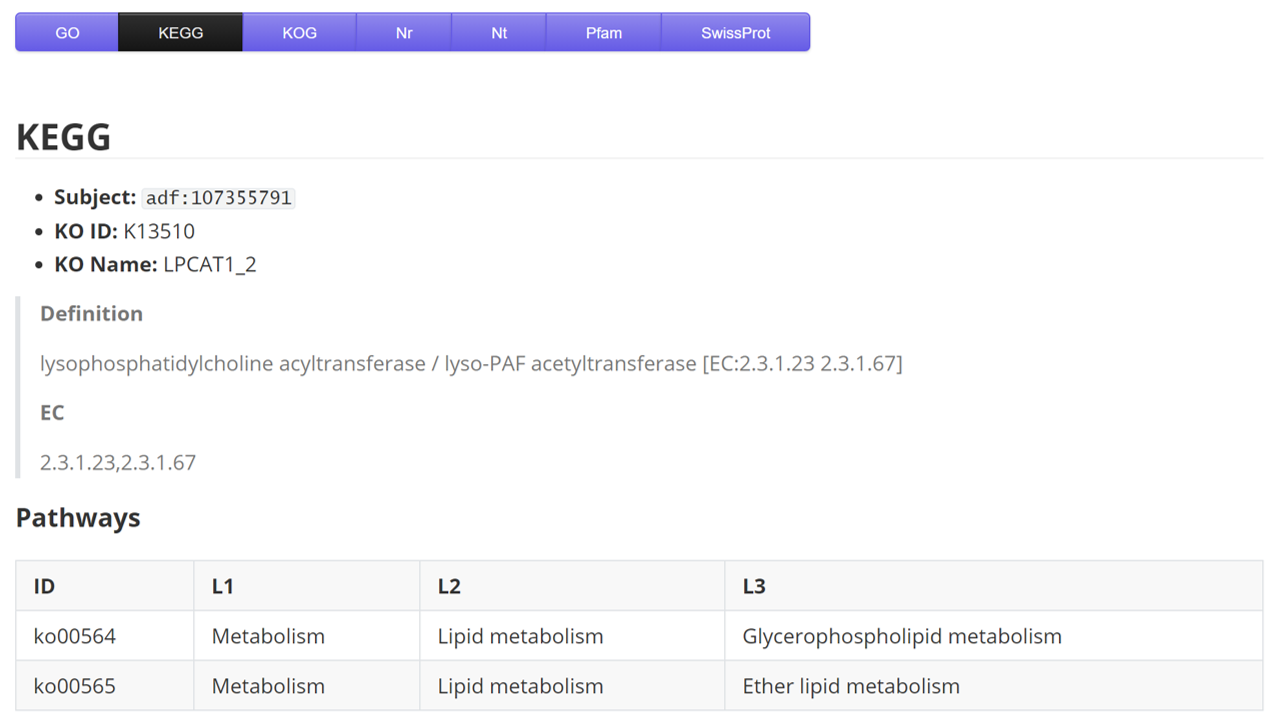
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...HQ_Formosa_2_transcript11807/f44p0/2477 |
| subject | target label | nve:NEMVE_v1g190252 |
| ko_id | KO ID | K03083 |
| ko_name | KO name | GSK3B |
| definition | KO description | glycogen synthase kinase 3 beta |
| ec | - | 2.7.11.26 |
| pathway | KO pathway | ko04012; Environmental Information Process... |
| species | coral species | Acropora muricata |
KOG
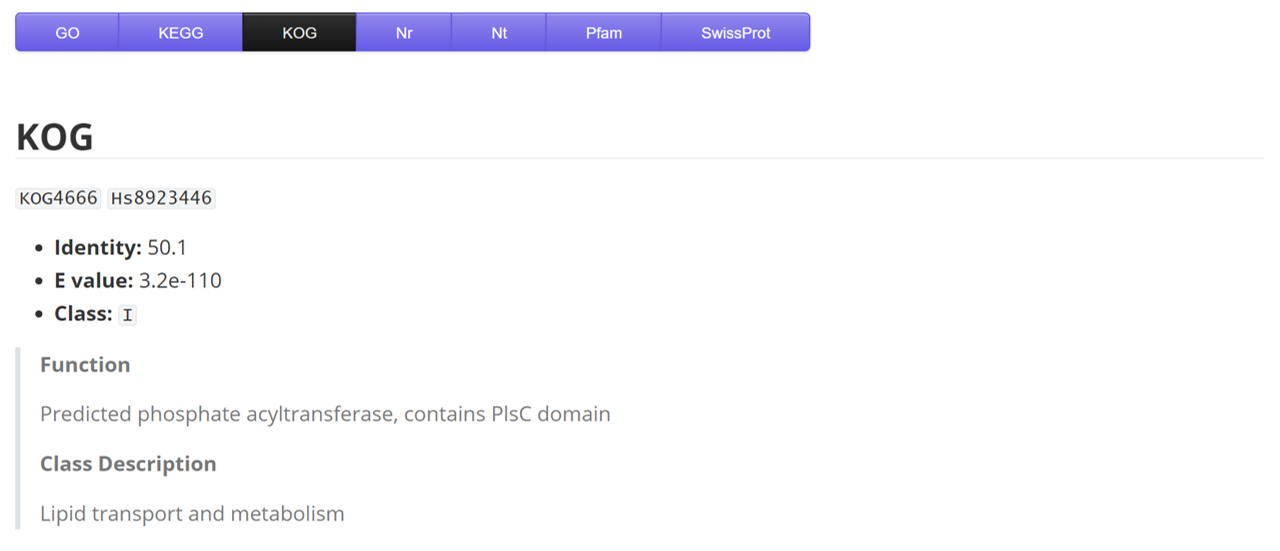
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...HQ_SH_981_transcript6218/f4p0/2707 |
| identity | - | 83.7 |
| E_value | - | 4.40E-160 |
| kog_gene | KOG gene ID | Hs19924133 |
| kog_id | KOG number | KOG1433 |
| function | function description | DNA repair protein RAD51/RHP55 |
| class | KOG classification | L |
| description | KOG class description | Replication, recombination and repair |
| species | coral species | Pocillopora verrucosa |
Nr
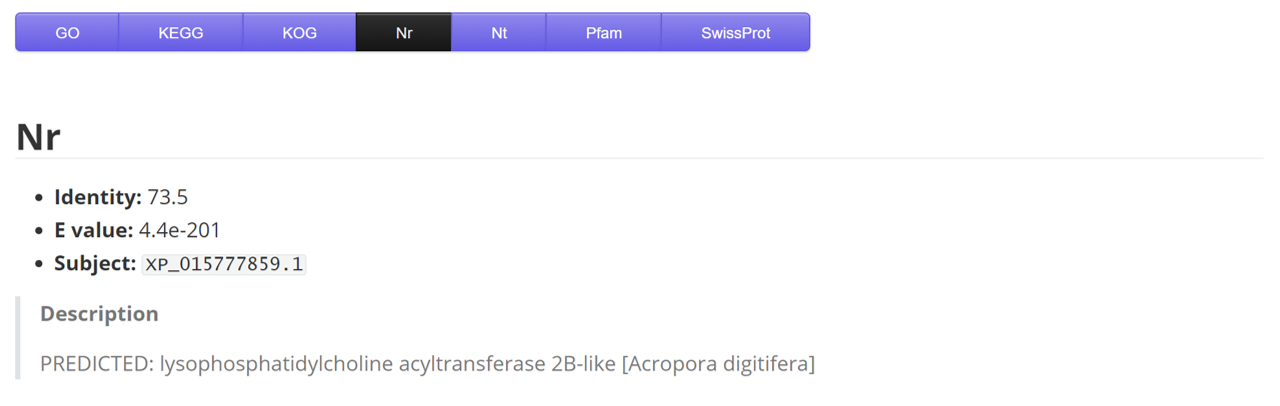
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...HQ_Formosa_2_transcript13610/f3p0/2327 |
| identity | - | 87.5 |
| E_value | - | 7.20E-168 |
| subject | target label | XP_001627328.1 |
| description | - | predicted protein [Nematostella vectensis] |
| species | coral species | Acropora muricata |
Nt
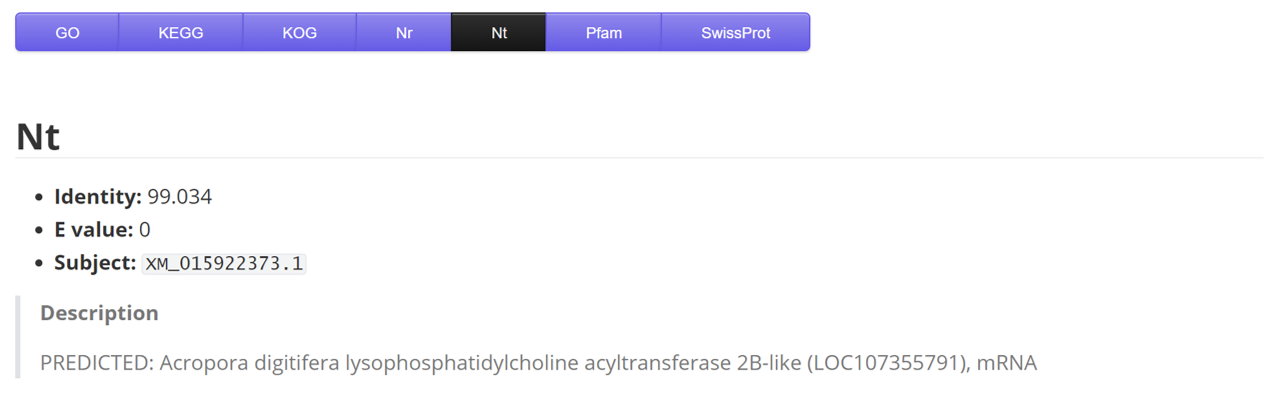
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...HQ_SH_FW_transcript9634/f2p0/2180 |
| identity | - | 78.743 |
| E_value | - | 2.52E-93 |
| subject | target label | XM_008286923.1 |
| description | - | Stegastes partitus rho-related GTP-bind... |
| species | coral species | Montipora capricornis |
Pfam
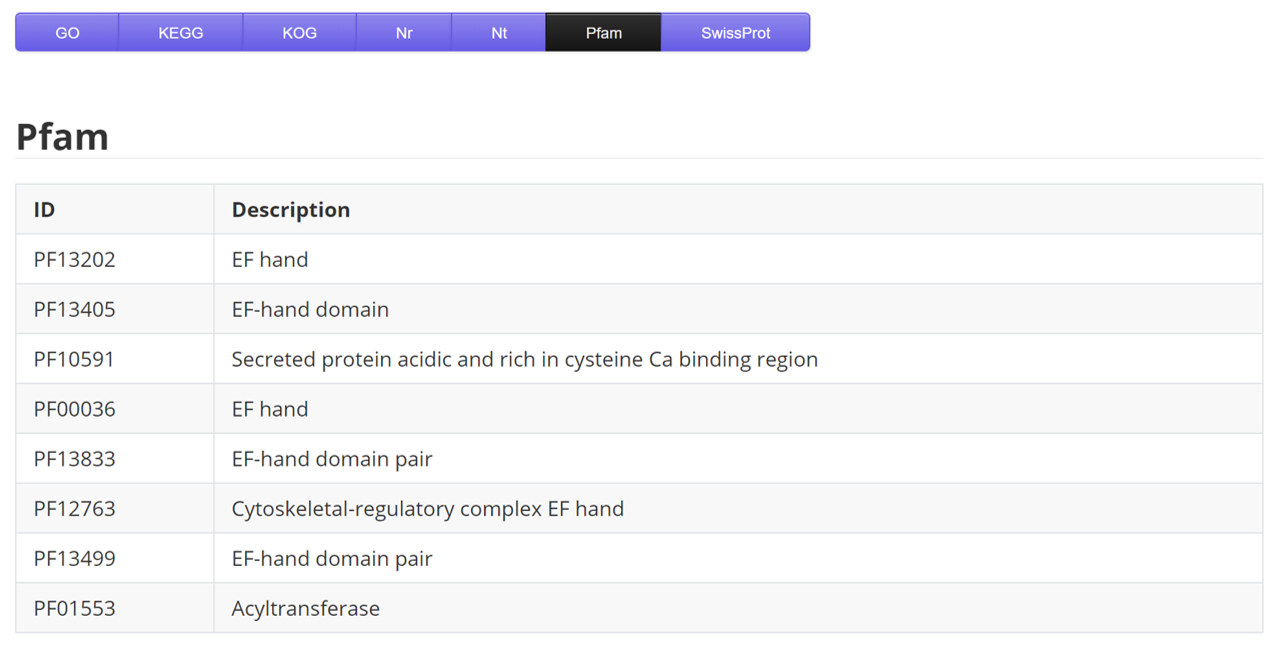
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...HQ_SH_WP_transcript19522/f3p0/1726 |
| id | PFAM ID | PF06293 |
| description | PFAM description | Lipopolysaccharide kinase (Kdo/WaaP) family |
| species | coral species | Montipora foliosa |
SwissProt
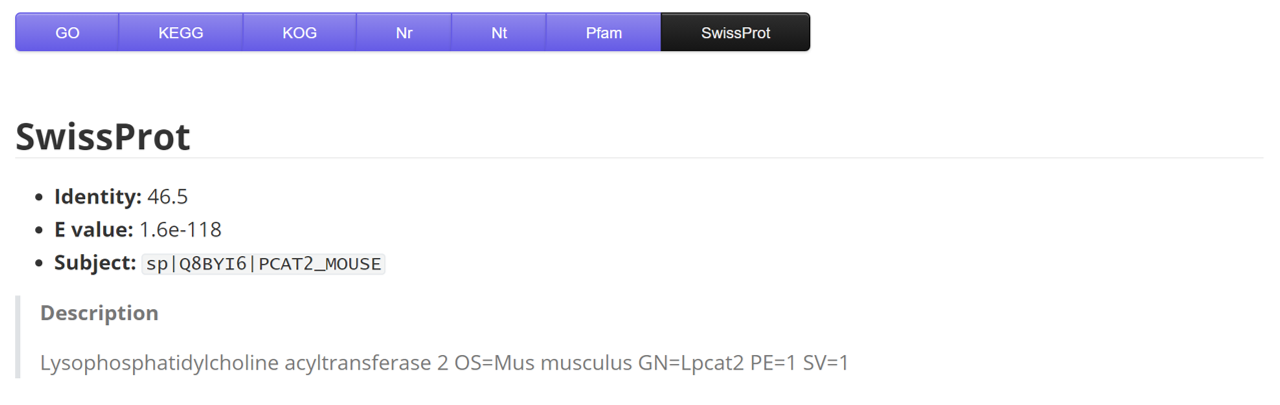
| Name | Description | Sample |
|---|---|---|
| gene | gene/transcript name | ...HQ_SH_WP_transcript22309/f13p0/1307 |
| identity | - | 88.5 |
| E_value | - | 1.50E-196 |
| subject | target label | sp|P62335|PRS10_ICTTR |
| description | - | 26S protease regulatory subunit... |
| species | coral species | Montipora foliosa |
1.3. Other data
2. WEB SERVICE
Tool module development is carried out simultaneously with data curation.
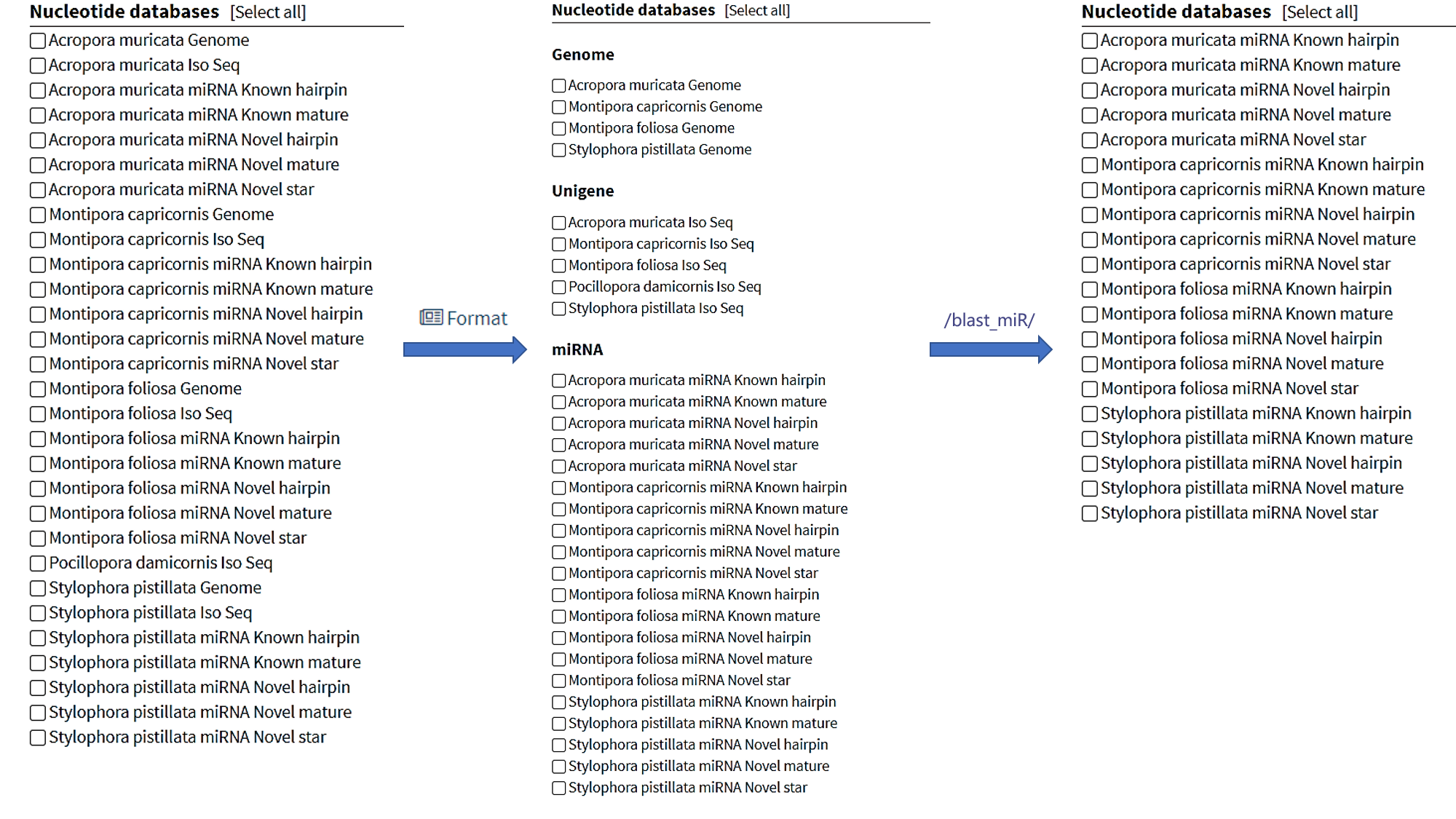
Optimized from Sequenceserver, it provides a more user-friendly BLAST interface. Click the "Format" button in the upper right corner to display data lists ordered by type, where sub-titles offer links to BLAST service for specific data.
Tips: Genomes have not been uploaded yet.
It is customized by GIVE to help gaining deeper insights into multi-omics data.
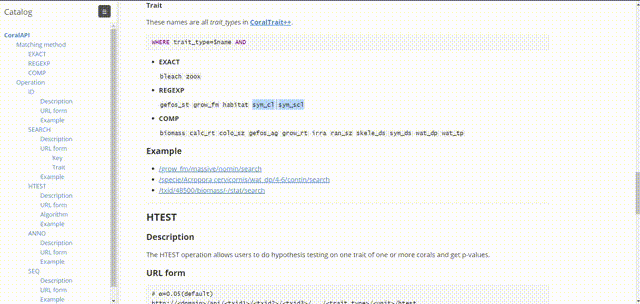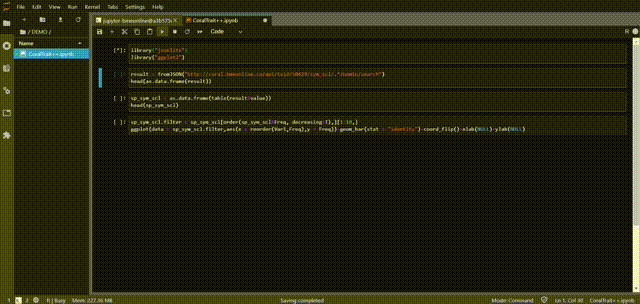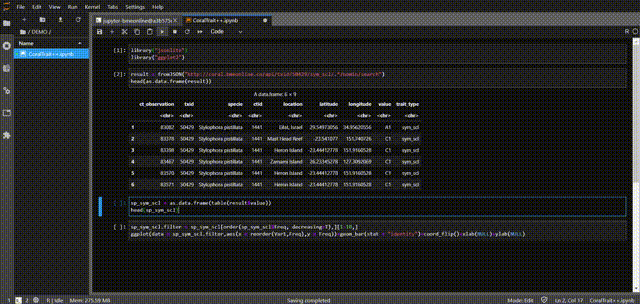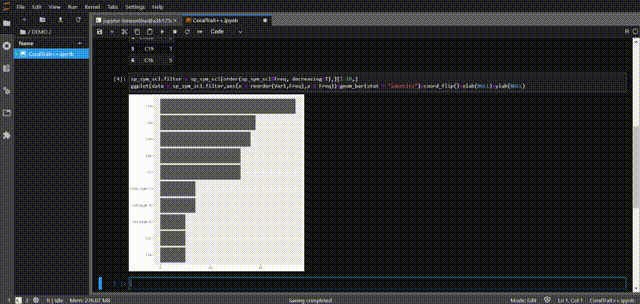
It offers users capability of acquiring data in-batch through URLs.
3. SUB-DATABASE
Two miRNA-centered sub-databases are built based on sequences and annotations from ▲CoralBioinfo.
It turns out to be the first database demonstrating reef-building coral miRNAs and their 3'UTR targeting sites.
It publishes a novel C1-clade Symbiodiniaceae miRNA set identified in four coral species.
4. INCLUSION CRITERIA
Contribution requests are always welcome, while they are ought to meet the basic requirement that all raw data must have mirrors on main public bioinformatics database such as NCBI. Please contact 📧Dr. Yunchi Zhu to offer your data after reading the following rules.
4.1. Genome
CoralBioinfo accepts reef-building coral genomes assembled to the chromosome level. The following data are required to be provided:
- Raw genome URLs on public database
- Assembled sequence (.fasta)
- Annotation (.gpd/.gff/.gtf)
CoralSym accepts C-clade Symbiodiniaceae genomes. The following data are required to be provided:
- Raw data URLs on public database
- Assembled sequence (.fasta/.fastq)
- Annotation (.gpd/.gff/.gtf)
Other genome-related data, which can be embedded into GIVE-customized genome browsers, are welcome as well.
4.2. Transcript
CoralBioinfo accepts reef-building coral full-length transcripts. Each full-length sample must be coupled with at least 3 NGS samples for correction and down-stream analysis such as DEG. The following data are required to be provided:
- Raw data URLs on public database (1 full-length sample against 3 NGS samples)
- Unigene sequence (.fasta)
- ▲Formatted annotation from at least 5 databases
4.3. Small RNA
CoralmiR accepts reef-building coral miRNAs. Submitted data format should be strictly in line with table structure of coralmir.info and coralmir.pair. In addition, the following data are required to be provided:
- Raw data URLs on public database
- miRNA-precursor structure (.ps/.pdf)
CoralSym accepts C-clade Symbiodiniaceae miRNAs. Before contribution, corresponding genomes need to be uploaded according to 4.1. Data format should be strictly in line with table structure of coralsym.info and coralsym.pair. In addition, the following data are required to be provided:
- Raw data URLs on public database
- miRNA-precursor structure (.ps/.pdf)

