Structure
Model Confidence
| █ | Very high (pLDDT > 90) |
█ | Confident (90 > pLDDT > 70) |
█ | Low (70 > pLDDT > 50) |
█ | Very low (pLDDT < 50) |
Predicted aligned error
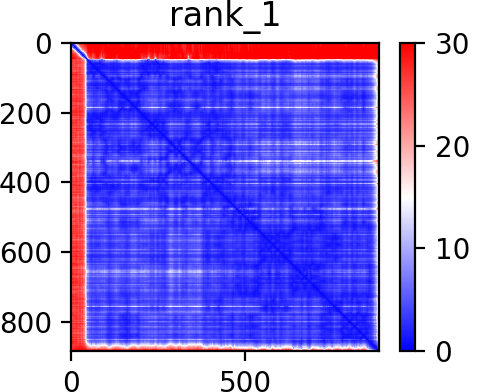
Homolog
| Database | Subject | Identity | E_value | Description |
|---|---|---|---|---|
| NR | XP_044162733.1 | 87.8 | 0.0 | dimethylglycine dehydrogenase, mitochondrial-like [Acropora millepora] |
| UniProt | A0A2B4SW28_STYPI | 84.7 | 0.0 | Dimethylglycine dehydrogenase, mitochondrial OS=Stylophora pistillata OX=50429 GN=Dmgdh PE=3 SV=1 |
Domain
| ID | description |
|---|---|
| IPR006076 | FAD dependent oxidoreductase |
| IPR006222 | Glycine cleavage T-protein-like, N-terminal |
| IPR013977 | Glycine cleavage T-protein, C-terminal barrel domain |
| IPR032503 | FAD dependent oxidoreductase, central domain |